News and Blogs
A Complete Guide to Doing an MRSA Swab Test
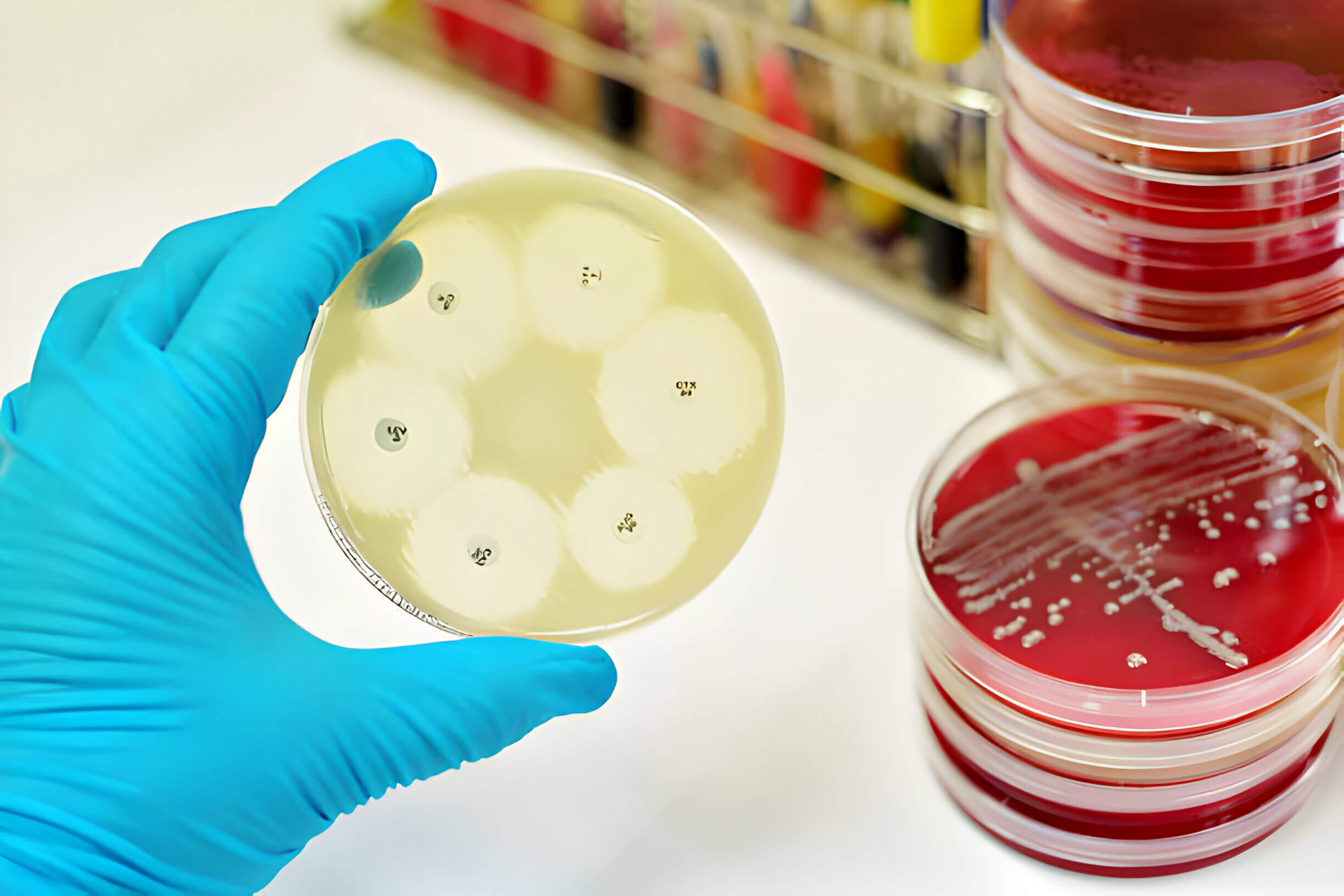
Methicillin-resistant Staphylococcus aureus (MRSA) is a specific type of staph bacteria often present on human skin or nostrils without causing harm. However, if these bacteria enter through an open wound, they can precipitate an infection. Most staph infections are minor and treatable with antibiotics. However, MRSA bacteria are resistant to common antibiotics, leading to their unchecked growth—a phenomenon known as antibiotic resistance. This resistance complicates treatments resulting in nearly 3 million infections and over 35,000 deaths annually in the United States.
While originally prevalent in hospitals, MRSA is now ubiquitous amongst healthy individuals. Its transmission can be via personal contact or interaction with contaminated objects. It is particularly prevalent in crowded environments like dormitories or locker rooms. Identification of MRSA is done through a test analyzing a sample from a wound, nostril, or other body fluid. If untreated, a MRSA infection can escalate to severe illness or death, but it can be remedied with potent antibiotics.
We will delve into how the test is conducted, and how doctors utilize this test in identifying and managing this challenging pathogen. Stay tuned for a comprehensive understanding of this process.
Instructions for Collecting an MRSA Swab Sample
Please follow these step-by-step instructions carefully when collecting an MRSA nasal, throat, and groin swab for testing.
Before Swab Collection:
- - Wash hands thoroughly with soap and water.
- - Assemble needed materials: swab packets, transport tubes, patient labels, biohazard bag.
Nasal Swabbing:
- 1. Open nasal swab packet and remove swab, handling the shaft only.
- 2. Insert swab tip into one nostril and rotate gently against nasal mucosa.
- 3. Repeat in second nostril using same swab.
- 4. Insert swab into transport tube and snap off shaft at break point.
Throat Swabbing:
- 1. Open throat swab packet and remove swab, handling the shaft only.
- 2. Insert swab tip into posterior pharynx and tonsillar areas and rotate gently.
- 3. Insert swab into transport tube and snap off shaft at break point.
Groin Swabbing:
- 1. Open groin swab packet and remove swab, handling the shaft only.
- 2. Swab skin folds in groin crease, rotating swab tip several times.
- 3. Insert swab into transport tube and snap off shaft at break point.
After Collection:
- - Label all transport tubes with patient identification stickers.
- - Place transport tubes into biohazard bag for processing.
- - Discard used swab packets appropriately.
MRSA Screening Test
Methicillin-resistant Staphylococcus aureus (MRSA) has become a major cause of healthcare-associated infections worldwide. MRSA refers to strains of Staphylococcus aureus that are resistant to all beta-lactam antibiotics, including penicillins, cephalosporins, and carbapenems. This resistance is mediated by the mecA gene which encodes an altered penicillin binding protein, PBP2a, that has low affinity for these antibiotics, allowing cell wall synthesis even in the presence of beta-lactams.
Accurate detection of MRSA in the clinical laboratory setting is critical for selecting appropriate antibiotic therapy and instituting effective infection control measures. However, determining methicillin resistance can be challenging due to the potential for heteroresistance, whereby subpopulations with varying susceptibility may coexist within an isolate. Careful testing methodology following standard protocols is therefore essential for the reliable phenotypic identification of MRSA strains.
Testing Theory
Accurately detecting methicillin resistance in Staphylococcus aureus can be difficult due to the heterogeneity that may exist within a bacterial culture. This phenomenon, known as heteroresistance, occurs when there is a mixed population of both resistant and susceptible subpopulations of cells coexisting within the same isolate.
The methicillin-resistant subpopulation has a slower growth rate compared to the susceptible cells. If testing conditions are not optimized, the faster-growing susceptible cells can outcompete the resistant ones, leading to false negative results. Several factors can contribute to this testing challenge:
- - Incubation Temperature: Resistant cells grow better at temperatures between 33-35°C. Higher temperatures favor growth of susceptible cells. Testing protocols must adhere to 33-35°C incubation for reliable detection.
- - Inducer Choice: Cefoxitin is a better inducer of mecA expression compared to oxacillin. It serves as an optimal surrogate marker and should be included in testing procedures.
- - Incubation Time: A full 24 hours incubation allows for adequate growth of slower-growing resistant subpopulations. Shortened incubation times risk missing the expression of resistance.
Full MRSA Detection Experiment Protocol
- 1. Isolate Pure Colonies: Obtain well-isolated S. aureus colonies from a culture plate to prepare a standardized cell suspension for testing.
- 2. Prepare Cell Suspension: Make a suspension of the isolate adjusted to a 0.5 McFarland turbidity standard to approximate a uniform inoculum density for testing.
- 3. Inoculate Test Plate: Inoculate the standardized suspension onto Mueller-Hinton agar test plates containing a cefoxitin disk. Add 4% NaCl to the agar. Use sterile swabs to evenly distribute the inoculum across the entire surface of the plate.
- 4. Incubate Plates: Invert inoculated plates and incubate at 33–35°C for a full 24 hours to allow for growth of any heteroresistant subpopulations.
- 5. Assess Growth: Examine plates for the presence of a zone of inhibition surrounding the cefoxitin disk. No zone indicates methicillin resistance (MRSA). Measure zone diameter if present.
- 6. Confirmation: Testing Perform confirmatory testing, such as mecA PCR, if phenotypic results are unclear.
Superior MRSA Screening with Mantacc Nasal Swabs
According to clinical research, nasal swabs demonstrate higher detection rates for MRSA screening compared to axillary or groin swabs. Optimized swabs enable superior performance in MRSA identification from nasal samples.
The Mantacc flocked nasal swabs are carefully designed to effectively collect microbiological nasal specimens such as MRSA. The soft, flocked tip features fine, flexible fibers to maximize sample collection from the nasal cavity while minimizing discomfort.
Additionally, we offer customized swab and kit solutions tailored to client requirements, including tubes with liquid or gel Amies transport media. Our flexible manufacturing capabilities allow us to engineer products suited for your exact MRSA testing procedures.
Mantacc aims to supply reliable tools to improve pathogen identification and streamline specimen collection workflows for clinicians. By providing optimized nasal swabs, it is our goal to assist healthcare staff in accurate first-time MRSA screening. Please contact our support team if you have any other questions. We look forward to helping advance your detection and surveillance needs.
Click to View → Mantacc MRSA Nasal Swabs
Related Posts
The Mechanism of MRSA Drug Resistance and Its Detection







