News and Blogs
Everything You Need to Know about Gene Sequencing

Gene sequencing technology is a method used to determine the sequence of DNA, from the initial Sanger sequencing, to the later Next Generation Sequencing (NGS), to the current single molecule sequencing technology. The development of sequencing technology has promoted the advancement of genomics, biomedical research, and clinical diagnosis. Here, we present you an overview of the trajectory of 3 generations of gene sequencing technology.
What is gene sequencing technology?
Gene sequencing technology, also known as the technology used for determining the sequence in nucleic acids.
Gene sequencing can analyze and map the complete sequence of a genome, pinpoint individual mutant genes, predict the likelihood of having multiple diseases, for early prevention and treatment.
Gene sequencing technology is one of the important methods for humans to explore the mystery of life. Initially, gene sequencing was only used in scientific research, serving as an important tool in genetics and molecular biology.
However, with the development of sequencing technology, through the decoding of genetic information and the construction of genomic databases, not only can humans peek at the code of life, but also detect and even intervene in human diseases at the genetic level.
Believe that under the guidance of gene sequencing technology, the diagnosis and treatment of genetic diseases, personalized precision medicine and other practices can work more efficiently. In the future, gene sequencing technology will have a significant impact on human health.
The development history of sequencing technology
In 1977, Sanger and Gilbert proposed the dideoxy chain termination method and the chemical degradation method respectively, marking the birth of the first generation of sequencing technology.
The first generation of sequencing has the advantages of long read length and high accuracy. However, it also has drawbacks such as high sequencing cost, long time consumption, and low throughput, which makes it unable to meet the demand for large-scale gene sequencing.
Therefore, people started to explore new and more efficient sequencing technologies.
In 1996, Ronaghi and Uhlen established Pyrosequencing, which, compared with the first-generation sequencing technology, sequences as it synthesizes. Its most notable features are high throughput and automation, so the second-generation sequencing is also called high-throughput sequencing.
In 2005, 454 Life Sciences company launched the Genome Sequencer 20 sequencing system based on the principle of pyrosequencing, becoming the pioneer of the second generation sequencing.
In 2006-2007, Illumina company and Life Technologies company successively launched Solexa high-throughput sequencing system and SOLiD high-throughput sequencing system.
In 2009, the third generation sequencing, represented by real-time sequencing at the molecular level and nanopore technology, emerged.The third-generation sequencing features long read lengths and single molecule sequencing. However, due to the high error rate of the current third-generation sequencing that has yet to be effectively addressed, there is still quite a long way to go to clinical application.
From 2010 to the present, various high-throughput sequencing technologies have developed rapidly and gradually matured. With the continuous development and integration of biological science, physics, materials science and other disciplines, future sequencing technology will certainly advance toward being more precise, more microscopic, higher throughput, and cheaper.
First generation sequencing (1stGS)
Basic Principle of Sanger's Dideoxy Chain Termination Method
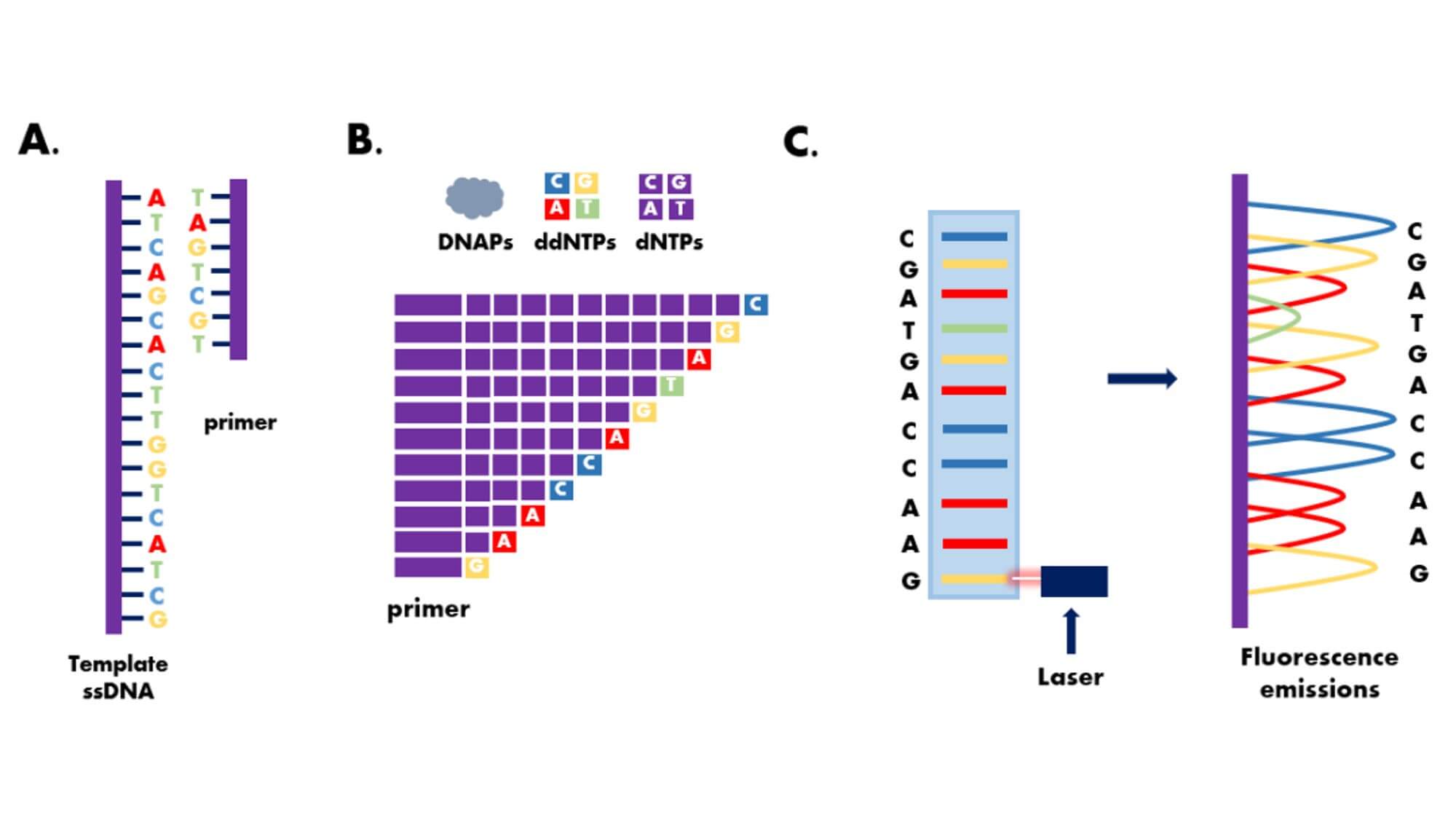
The Sanger dideoxy chain termination method is the most classic one in the first generation sequencing technology.It cleverly uses the principle of DNA replication, using ddNTP to partially replace conventional dNTP as the substrate for DNA synthesis.During DNA synthesis, once a ddNTP is incorporated into the synthesizing DNA chain, because the 3'-carbon atom of the deoxyribose of ddNTP lacks a hydroxyl group, it cannot form a 3',5'-phosphodiester bond with the phosphate group of the next nucleotide, thus causing the elongating DNA chain to terminate at this ddNTP site.
Experimental Steps:
- 1. Amplify the DNA fragment to be sequenced by PCR to obtain sufficient DNA templates for sequencing.The obtained DNA templates need to be purified, making sure to remove all impurities, including DNA fragments, proteins, RNA, etc.
- 2. Design some specific primers according to the characteristics of the sequence to be sequenced and experimental requirements.Generally, the length of universal primers should be 15~30bp.
- 3. Synthesis of labels.The label is a substance indicating whether the sequencing reaction is successful, usually a fluorescent dye or radioactive isotope.
- 4. Mix substances such as the DNA template, primers, DNA polymerase, and labels together for the sequencing reaction.During the reaction, the DNA polymerase synthesizes new DNA chains based on the base sequence in the DNA template and combines labels into the newly synthesized DNA chain.
- 5. Electrophoretically separate the reaction products and determine the order of bases in the DNA sequence based on the different fluorescent signals or the presence of radioactive isotopes of the labels.On the basis of the Sanger sequencing method, the use of fluorescent signal receivers and computer signal analysis systems instead of the autoradiography technology, and the use of fluorescent label instead of single radioactive isotope labels (such as 32P or 35S), has opened the door to the automation of DNA sequencing technology.
Pros and Cons of Sanger Sequencing:
Pros:
- 1. The Sanger method directly sequences DNA molecules, suitable for validation sequencing of known sequences, library screening, clone identification, and PCR resequencing, and so on.
- 2. Its biggest advantage is high reading speed, high accuracy, and relatively low cost. Compared to chemical degradation sequencing, the sequencing effect will not be affected in regions rich in G-C.
Cons:
- 1. A primers with a known sequence must be designed for sequencing, for unknown sequences cloning needs to be constructed before sequencing can be done, making it difficult to achieve large-scale sequencing at the genome level.
- 2. Determining the base sequence requires a large number of identical DNA copies.

Second generation sequencing (2ndGS)
With the completion of the Human Genome Project, which spanned 13 years and cost nearly $300 million, life science entered the epochal era of functional genomics.
People began to hope to find the exact mechanism of disease occurrence in the gene map and implement precise medical plans.
Although the first-generation sequencing technology has advantages such as long read length and high accuracy, its high sequencing cost, time-consuming, and low throughput deficiencies make it unable to meet the needs of large-scale sequencing.
In 1996, Ronaghi and Uhlen established pyrosequencing. In 2005, the 454 Life Sciences company launched the Genome Sequencer 20 system based on the principle of pyrosequencing.
This is a milestone event in the history of sequencing, it changed the scale of sequencing and became the forerunner of the second-generation high-throughput sequencing.
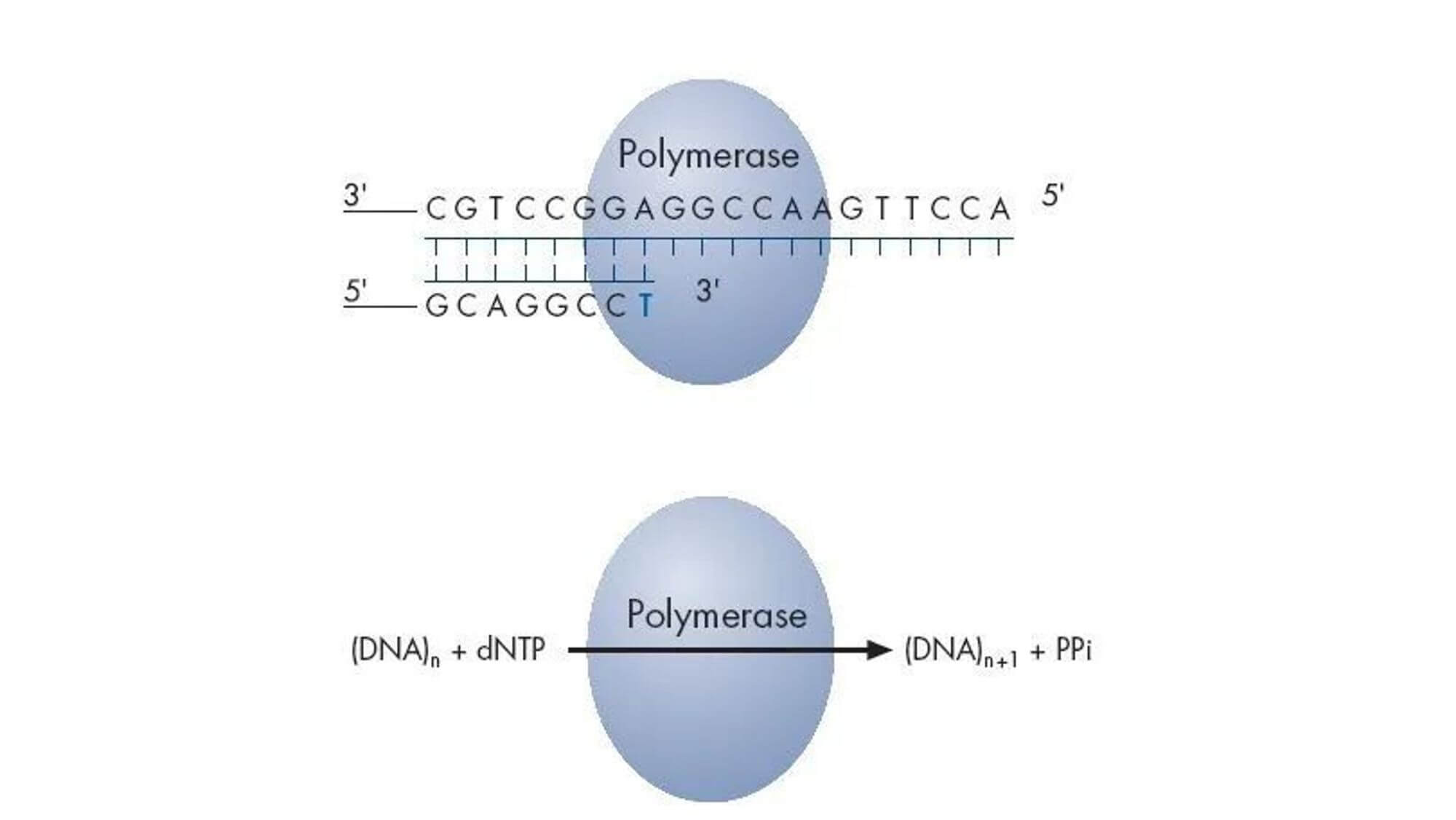
The core concept of second-generation sequencing technology is sequencing while synthesizing, its most notable features are high throughput and automation.
Unliked the Sanger sequencing technology, which performs individual reactions after cloning the template, the second-generation sequencing technology breaks up the template DNA into small fragments and amplifies the library through bridge PCR (or emulsion PCR), while sequencing hundreds of thousands to millions of DNA templates at the same time.
The emergence of the second-generation sequencing technology has made deep sequencing of a species' genome and transcriptome no longer distant, it can maintain a high degree of accuracy, while lowering the cost of sequencing and increasing the speed of sequencing.
Taking the human genome as 3Gb, using the first-generation sequencing technology, about 62500 times of sequencing is needed to complete the human genome sequencing. Counting each reaction as 2 hours, assuming 10 times of sequencing per day and working 7 days per week, the whole process would take about 17 years, while using high-throughput sequencing technology, the human genome sequencing can be completed in just 1 week.
Principle of Pyrosequencing & 454 Sequencing System
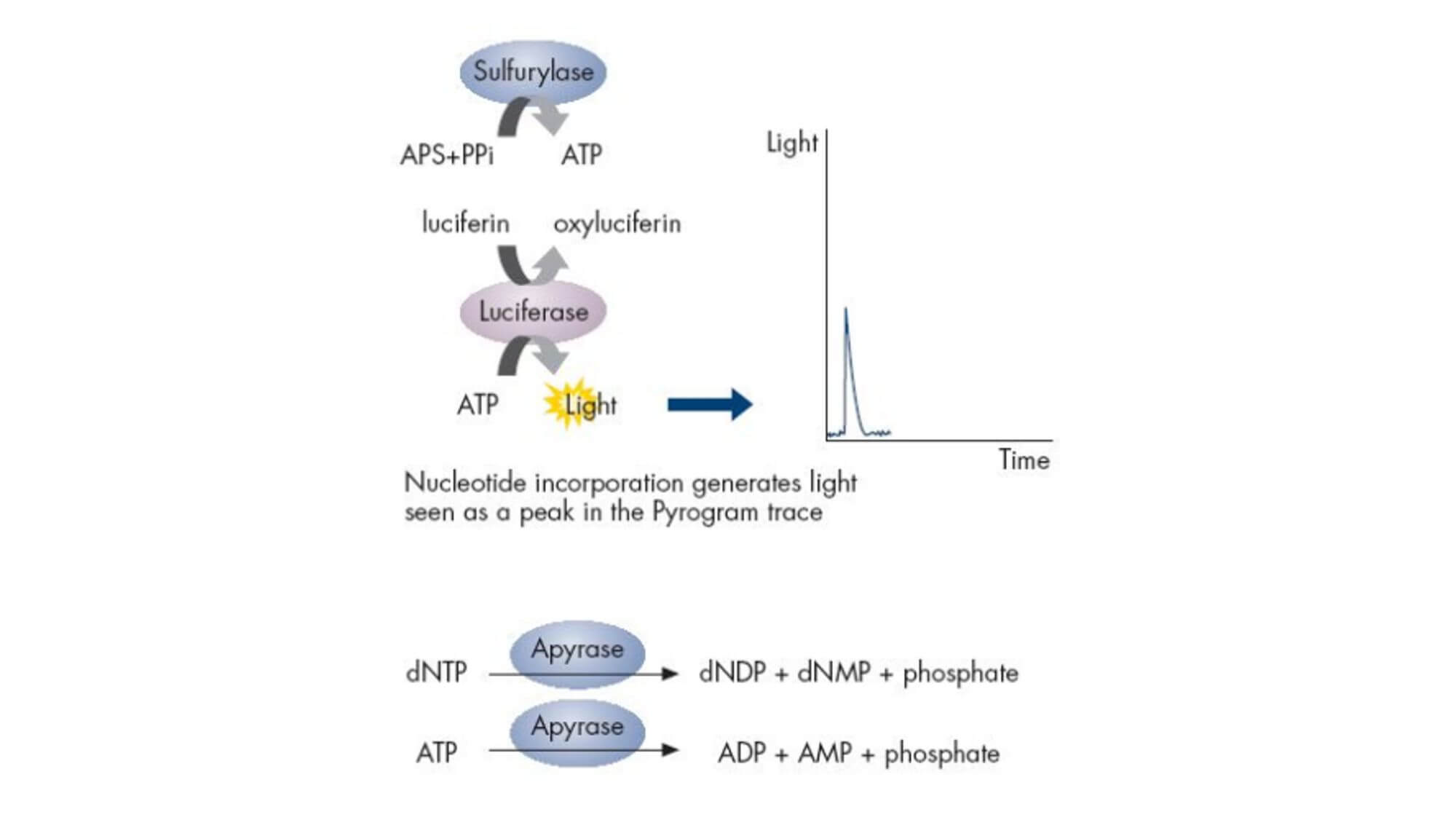
Pyrosequencing is a novel enzyme cascade chemiluminescence sequencing technology catalyzed by DNA Polymerase, ATP Sulfurylase, Luciferase, and Apyrase. By performing real-time detection on the biological light signal released during DNA synthesis, it paved the way for sequencing while synthesizing.
Experimental Principle:
The reaction substrates are 5'-adenosine phosphosulfate (APS) and luciferin. In each round of sequencing, only one type of deoxyribonucleotide triphosphate (dNTP) is added to the reaction system. If it exactly matches the next base of the DNA template, it will be added to the 3' end of the sequencing primer under the action of DNA polymerase, simultaneously releasing a molecule of pyrophosphate (PPi). Under the catalysis of ATP Sulfurylase, the produced PPi can bind with APS to form ATP, and under the catalysis of Luciferase, the generated ATP can bind with luciferin to form oxyluciferin, simultaneously producing visible light. A specific detection peak can be obtained through a weak light detection device and processing software, and the height of the peak is directly proportional to the matched base. If the added dNTP cannot pair with the next base of the DNA template, the above reaction will not occur, and there will be no detection peak.
ATP and unincorporated dNTPs are degraded by pyrophosphatase, starting a new cycle.
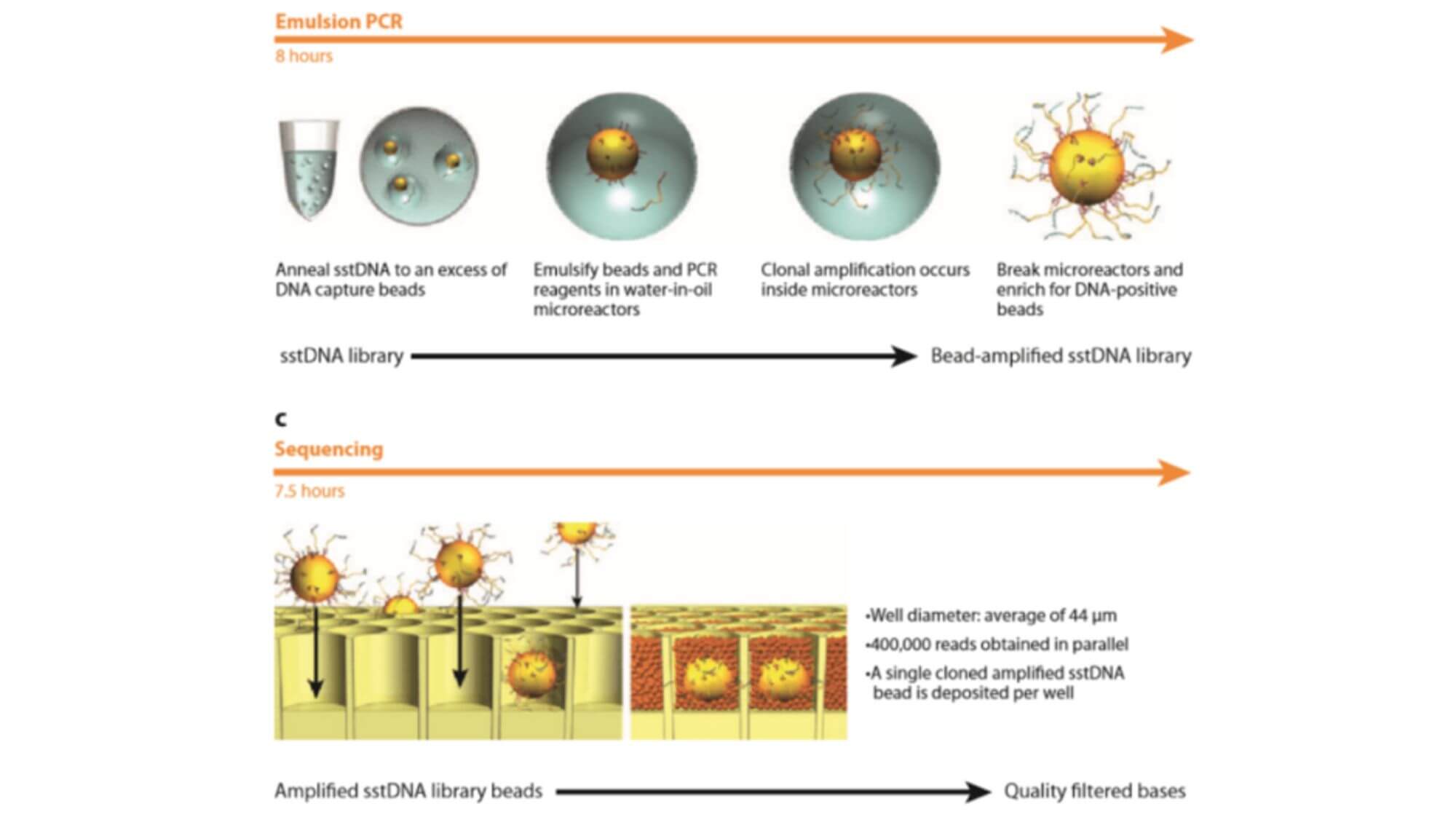
In 2005, the 454 Life Sciences company combined pyrosequencing technology with emulsion PCR and optical fibre chip technology to launch the Genome Sequencer 20 high-throughput sequencing system. This initiated large-scale parallel pyrosequencing, achieving high throughput in the sequencing process.
Emulsion PCR experimental principle:
Emulsion PCR is the encapsulation of the aqueous phase by the oil phase, and using the encapsulation structure as a microreactor for PCR amplification. The biggest feature of emulsion PCR is that it can form a large number of independent reaction spaces for PCR amplification.
The process of "oil encapsulates water":
- 1. Before the PCR reaction, the aqueous solution containing the template, dNTP, primer, and DNA polymerase is injected onto the surface of high-speed rotating mineral oil. The aqueous solution instantly forms countless water droplets encapsulated by mineral oil.
- 2. The "oil encapsulates water" package contains magnetic beads, the surface of which contains DNA sequences complementary to the adapters, allowing single-stranded DNA sequences to bind specifically to the magnetic beads.
- 3. The magnetic bead contains the reagents required for the PCR reaction, ensuring that each fragment bound to the bead can be independently amplified by PCR, and that the amplified product can still be bound to the bead. Beads carrying amplified DNA fragments are placed in a PTP plate for sequencing.
In 2007, after being acquired by Roche, 454 Life Sciences company launched the second-generation sequencing system - Genome Sequencer FLX System, which has an even better performance. The long-read exceeds 400bp, providing about one million sequences in 10 hours, with 400 to 600 million bases information, and an accuracy exceeding 99%.
The 454 high-throughput sequencing system has obvious advantages in read length, making subsequent assembly work more efficient and accurate. It is the ideal choice for de novo genome sequencing, transcriptome analysis, and genome structure analysis applications.However, since it uses the pyrosequencing principle to detect instantaneous luminescence, this limits its greater throughput, and the detection of homopolymers (sequences where the same base is consecutively present a few times) is not accurate enough, the longer the homopolymer, the greater the potential error.
In addition, compared to other high-throughput sequencing platforms, the cost of pyrosequencing is much higher, and it didn't lead with its early-adopter advantage in intense market competition.
In 2013, Roche officially announced the closure of the 454 sequencing business.
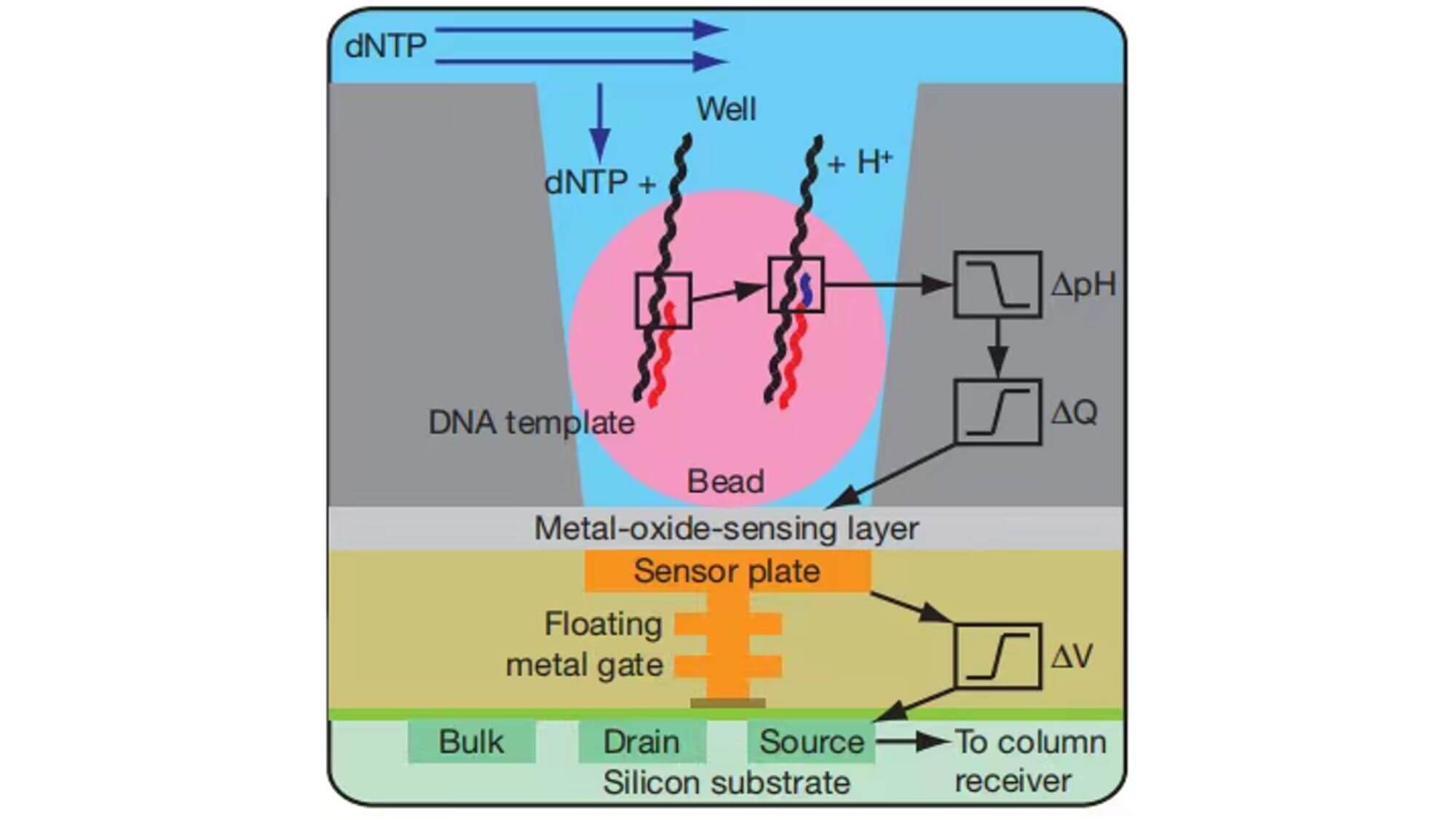
Ion Torrent Sequencing System
In 2007, after leaving LifeSciences Company, Rothberg immediately founded Ion Torrent Company and developed a revolutionary new high-throughput sequencing platform based on a semiconductor chip. The Ion Torrent sequencing system is the first high-throughput sequencing platform with no optical sensor. Ion Torrent sequencing uses a semiconductor chip as a carrier, and detects the pH change caused by the release of H+ during DNA chain synthesis, transforming chemical signals into electrical signals to obtain base information, implementing the sequencing while synthesizing technology.
Sequencing process:
- 1. The Ion Torrent sequencing system also uses emulsion PCR technology. The sequencing reaction takes place on an Ion Torrent chip (a high-density semiconductor chip full of small holes). Each small hole can only accommodate one sequencing bead. The bottom of the hole is a pH-sensitive field-effect transistor sensor, which can detect changes in pH within the hole and convert chemical signals into digital information.
- 2. The prepared bead suspension is injected from the chip's entrance, and the chip is centrifuged to better trap individual sequencing beads in individual small holes. The more holes on the chip, the greater the sequencing throughput.
- 3. Solutions containing A, T, C, G four different dNTPs flow over the chip separately. If the added dNTP successfully pairs with a base on the DNA chain, it will release one H+ ion, leading to a change in the pH value inside the small hole, which is detected and recorded by the sensor at the bottom of the chip. When consecutive identical dNTPs bind to the DNA chain, the same number of H+ will be released, and the signal recorded by the sensor will also double accordingly. If the added dNTP cannot match the base on the DNA chain, no reaction will occur, and there will be no change in the pH value inside the hole, therefore, no base will be recorded. The unbound dNTP and the remaining primer and enzymes are washed away, then the next dNTP solution is added, and the sequencing repeats in this manner.
- In the early stage of sequencing, A, C, G, T, these four bases' detected pH changes are used to determine the signal strength baseline of the entire bead. With the standard signal strength, the signal measured afterwards is compared with these four signal strengths. If it is 1 times the strength, it indicates that there is one base, if it is 2 times the strength, it indicates that there are two identical bases, and so on. However, due to the possible deviation of the sensor's sensing of the current, there could be a bias in determining the number of consecutive bases.
In 2010, after acquiring Ion Torrent, Life Technologies quickly launched the Ion PGM sequencer. This device, named the "Personal Genome Sequencer", is the world's first DNA decoder reliant on silicon transistors, capable of accurately reading 10 million genetic codes in 2 hours. Since there is no need for labeling, lasers, imaging equipment, etc., the price is much lower than other sequencers, with a sale price of only $50,000, it was commonly regarded as the smallest, cheapest genetic decoder on the market at that time. This economical and fast sequencer is instrumental for the popularization of sequencing technology and also brings hope to rapid clinical gene testing.
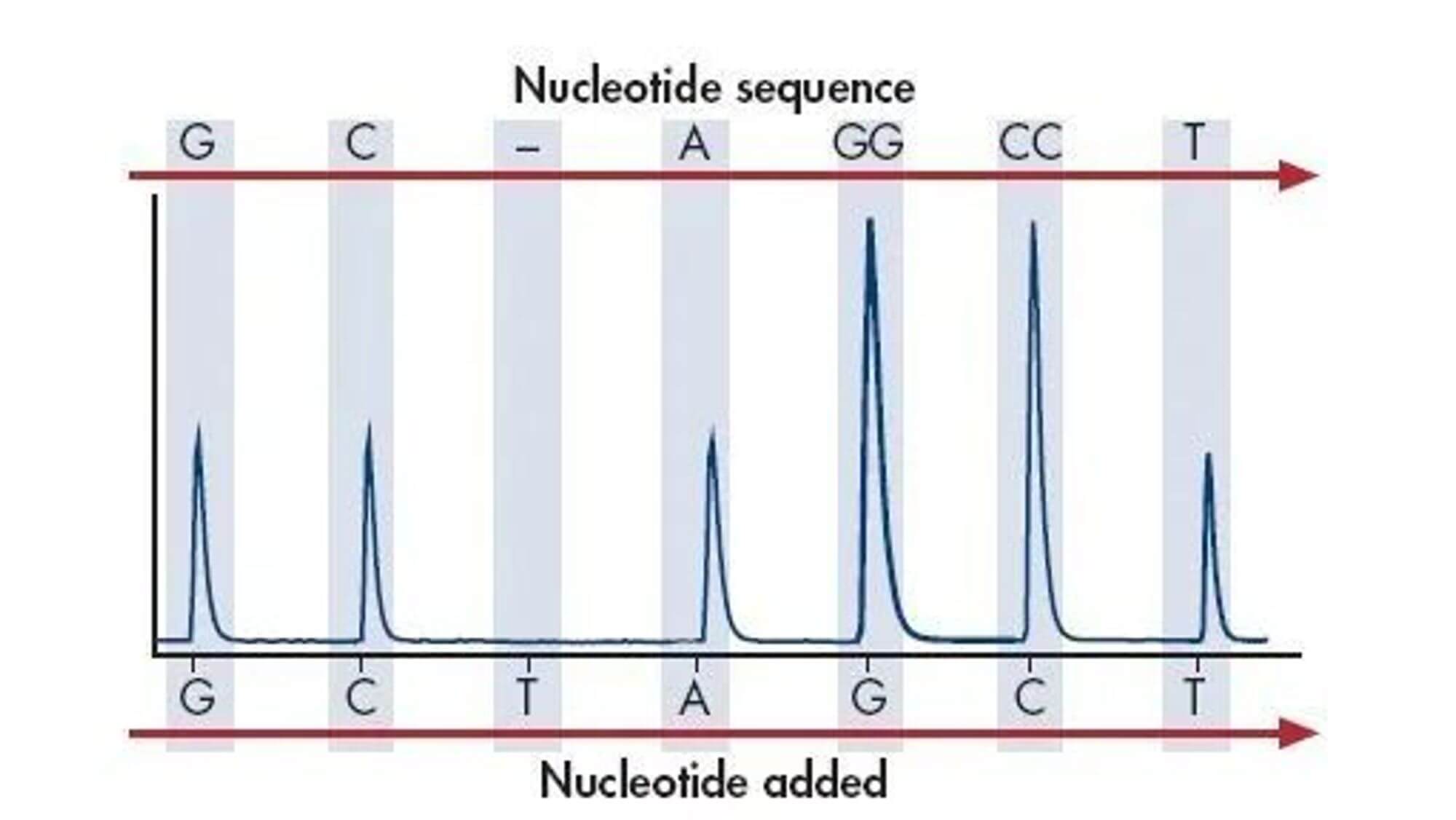
Solexa Sequencing System
In 2006, Solexa Company launched the Genome Analyzer.
In 2007, Illumina Company purchased Solexa at a high price and commercialized it. The Solexa sequencing system still uses sequencing while synthesizing as its basic design concept, and employs bridge PCR and reversible terminator as its core technologies.
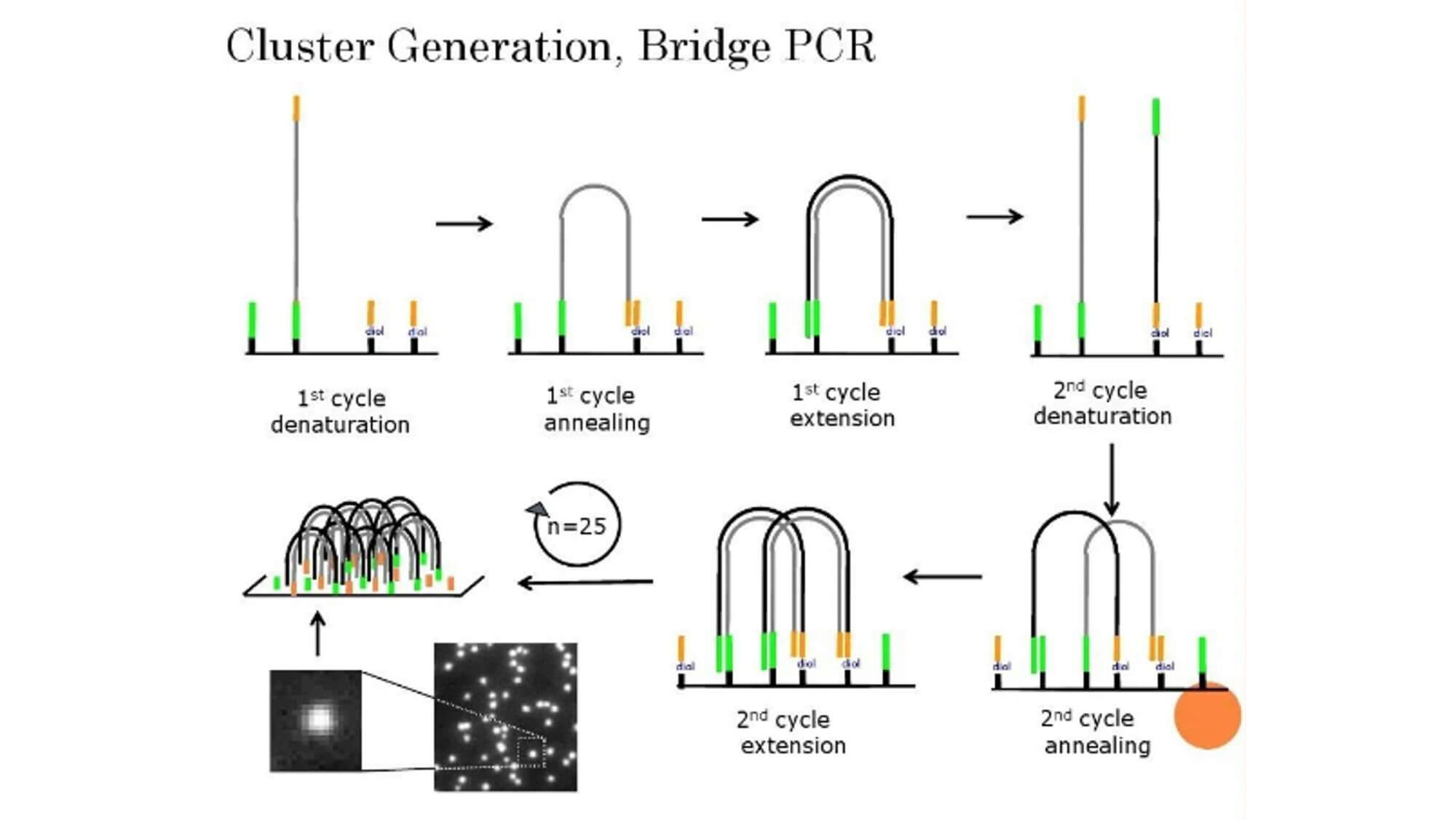
The basic principle of bridge PCR:
Bridge PCR is the process of fixing DNA fragments to a chip and then amplifying them with PCR. First, DNA fragments are mixed with primers, and then polymerase and dNTPs are added to amplify them. In the amplification process, DNA fragments will bind to the primers on the surface to form a bridge structure. This bridge structure can maintain the stability of the DNA fragments and allows for high-throughput sequencing on the surface.
Sequencing process:
-
The genome DNA is broken into small fragments of several hundred bases (or shorter), and adapters are added to both ends of the fragments.
-
The surface of the chip is connected with a layer of single-stranded primers. After the DNA fragment becomes single-stranded, it is "fixed" at one end on the chip through base complementarity with the primer on the chip surface.
The other end (5' or 3' end) randomly complements another primer nearby and is also "fixed", forming a "bridge". After repeating 30 rounds of amplification, the final result is approximately 1000 copies of monoclonal DNA clusters. After the DNA clusters are formed, the amplification products are linearized. Sequencing primers subsequently hybridize on the common sequence on one side of the target area to carry out the sequencing while synthesizing reaction.
-
The Genome Analyzer system uses the principle of sequencing while synthesizing.Modified DNA polymerase and 4 types of dNTPs (each type of dNTP is linked with a fluorescent group) are added. These dNTPs are "reversible terminators" because the 3'-OH terminus carries a chemically cleavable segment that only allows a single dNTP to be incorporated in each cycle.
At this point, a laser scans the surface of the reaction plate to read the type of dNTP polymerized in the first round of reaction for each template sequence. Afterwards, the remaining dNTPs, DNA polymerases and fluorescent groups are removed, and the stickiness of the 3' end is restored to continue to polymerize the second dNTP.
This process continues until each template sequence is fully polymerized into a double-strand. In this way, by counting the fluorescent signals collected in every round, we can learn the sequence of each template DNA fragment.
Since Solexa's technology can only add one dNTP at a time in the synthesis process, it effectively solves the accuracy issue of homopolymer (a series where the same base is consecutively repeated several times) detection.
Illumina platform has dominated the second-generation sequencing market, and Genome AnalyzerIIx and HiSeq high-throughput sequencers are the most widely used second-generation sequencers worldwide.
The NovaSeq series launched by Illumina in 2017 operates 70% faster than existing instruments, and can complete whole-genome sequencing in just 1 hour. It is considered to be the most powerful sequencer Illumina has launched to date, signaling the arrival of the $100 genome era.
Complete Genomics Sequencing System
Founded in 2005, Complete Genomics (CG) in the United States is the world's first life science company to provide human genome sequencing services. The CG company uniquely owns two sequencing-related technologies, the DNA nanoball (DNB) chip and the combinatorial probe anchor ligetion (cPAL), which offer 99.9998% sequencing accuracy at a low market price and thus possess significant competitive advantages.
The library construction in cPAL sequencing is called DNB, which uses Rolling Circle Amplification (RCA) to amplify DNA into a linear spiral structure. The advantage of this method of library construction is that all amplified templates are the original insert fragments. In this way, errors produced by PCR will not accumulate and will only affect the amplified sequence. In contrast, if an error occurs during the amplification in Illumina sequencing, the subsequent amplification will use this erroneous fragment as a template, leading to the accumulation of errors.
RCA amplification:
RCA uses a short circular oligonucleotide as a template, with dNTPs as a raw material, and generates a long repetitive single-strand DNA/RNA under the effect of a DNA/RNA polymerase.
Working Principle:
-
1. The template for rolling circle amplification must be circular. If linear genes are amplified, then a locking probe is needed. Both ends of the locking probe have sequences complementary to the target gene. After the locking probe recognizes the target gene and binds to it, it forms an incompletely closed circular oligonucleotide. Under the action of the ligase, it becomes a completely closed circular oligonucleotide. If the DNA is circular to begin with, this process is not necessary.
-
2. Linear amplification: The forward primer identifies the pair sequence of the circular template, synthesizing a repetitive linear single-strand DNA sequence under the action of Phi29 DNA polymerase. This single-strand DNA contains hundreds to thousands of repetitive template complementary segments.
The amplification products of RCA are a single-strand DNA that forms a linear spiral, which is referred to as the DNA nanoball. After the library is built, it is added to the sequencing chip. The sequencing chip has a DNB binding site, with one site binding one DNB. Then it proceeds with cPAL sequencing which is similar to SOLiD.
The process is that:
In each round of sequencing, an oligonucleotide anchor sequence that matches the adapter is added first, followed by a probe that contains different known bases and a fluorescent group.
Each probe only has one base carrying a fluorescent marker (the position of this fluorescent-marker base in the probe is determined by the sequencing position. For example, if you want to test the first base, then you only mark the first base of the probe. If you want to test the fifth base, then mark the fifth base of the probe).
In each round, only one probe can pair with the sequencing sequence. After pairing with the sequencing sequence, remove the other unpaired probes, then detect the fluorescent signal and obtain the sequence information. Then, all the binding probes and anchor sequences are removed to start the next round of sequencing.
Compared with Illumina's SBS sequencing, the advantage of this is that the next base does not depend on the previous base, so sequencing errors are more random.
The cPAL technology dramatically reduces the concentration of probes and enzymes. In addition, unlike sequencing while synthesizing, cPAL can read several bases at once in each cycle.
In this way, the consumption of sequencing reagents and imaging time are substantially reduced. Currently, the read length of this high-throughput sequencing platform is 28~ 100bp, which greatly reduces the operability of genome assembly and limits its application in structural variation research.
Summary
In general, while the second-generation sequencing technology meets the demand for throughput, due to its inherent technical limitations, the length of the single sequence read is 75~100bp. This forms the current technical bottleneck of high-throughput sequencing – high throughput results in shorter read length, and longer read length results in lower throughput.
The throughput determines the cost and duration of the sequencing, while the read length determines the difficulty of piecing together and restoring the real situation of the genome from the obtained DNA fragments.
We can imagine the assembly process as a puzzle game, with each piece of DNA sequence information representing a puzzle piece. The bigger each puzzle piece is, the easier it is to assemble into the original picture. This aptly explains why sequencing technologies continuously strive for larger fragments and longer read lengths while pursuing high throughput.
The existing second-generation sequencing technologies are identified through the collection of fluorescent signals, so library construction is required for amplification and reaction. This part is the most susceptible to human interference in the second-generation sequencing technology. Due to variations in the proficiency level of practitioners, even the same equipment can perform differently in different laboratories.
Moreover, using the amplified products as sequencing templates may result in errors during amplification, missing information (such as methylation), and sequence bias. This can lead to fragments with low copy numbers in the original sample being obscured after the amplification reaction; certain modification information in the original sequence may also be obliterated during the amplification process. Although researchers have made significant efforts in the development of software and algorithms, limitations in the analysis of second-generation sequencing data still exist.
Third generation sequencing (3rdGS)
The ideal sequencing technology is one that allows direct and accurate sequencing of the original DNA template without being limited by read length.
As early as the 1980s, researchers began to strive to achieve this goal. Although many attempts at this failed, single-molecule real-time sequencing technology and nanopore sequencing technology eventually made it possible to sequence single molecules with long read lengths, once again revolutionizing the field of sequencing.
Sequencing technologies characterized by unamplified single-molecule sequencing and long read lengths are referred to as third-generation sequencing technologies.
These technologies can read fragments as long as tens of thousands of bases in a single run, greatly reducing the difficulty of assembly, and more importantly, significantly reducing the number of gaps that could not be mapped in the past.
However, current third-generation sequencing technologies still have not found a good solution for their high error rates, and there is still a considerable distance before they can be practically applied in the clinic.
Pacific Biosciences SMRT Sequencing Technology
SMRT sequencing technology was proposed by Webb and Craighead, and further developed by Korlach, Turner, and Pacific Biosciences (PacBio), and was launched as the PacBio sequencing platform in 2009.
SMRT sequencing technology is based on single-molecule reading technology of nano-pores and can quickly complete sequence reading without amplification.
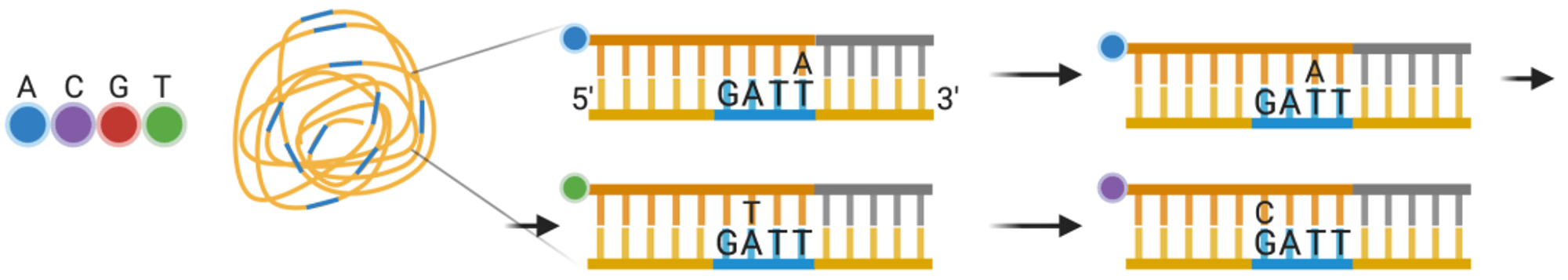
SMRT sequencing technology uses a specially made fluid unit (SMRT cell), which contains thousands of sequencing micro-pores (picolitre wells) — Zero-mode waveguide (ZMW) holes, which is one of the key points of SMRT technology.
It can distinguish the reaction signal from the strong fluorescence background of free dNTPs. Its basic principle is the same as that of Illumina, which is sequencing while synthesizing.
Sequencing process:
-
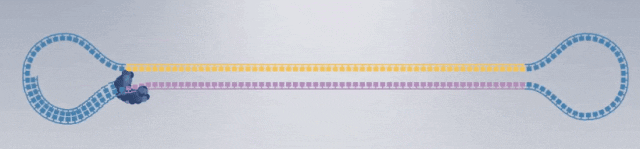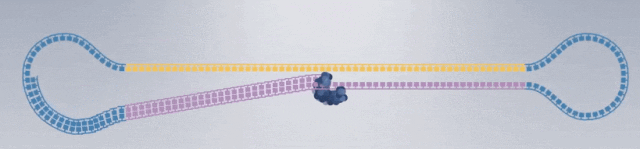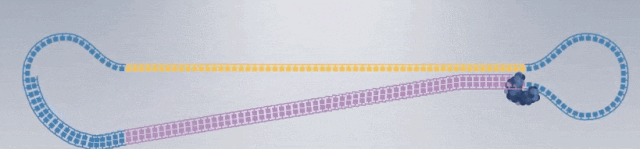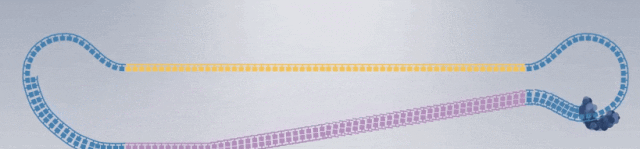
1. After extracting the DNA or RNA molecules from the sample, construct the following dumbbell-shaped molecular structure: Dumbbell-shaped molecular structures are constructed from all DNA fragments in the sample, forming a set called a library (SMRTbell Library), which will then be placed in the sequencing chip.
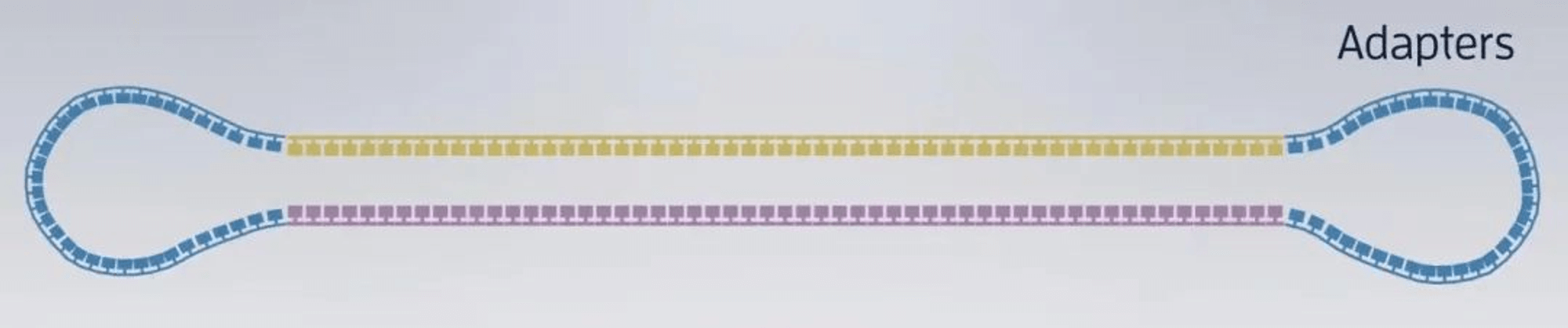
-
2. Taking RSII sequencing platform as an example, the sequencing chip (SMRT Cell) looks like this:
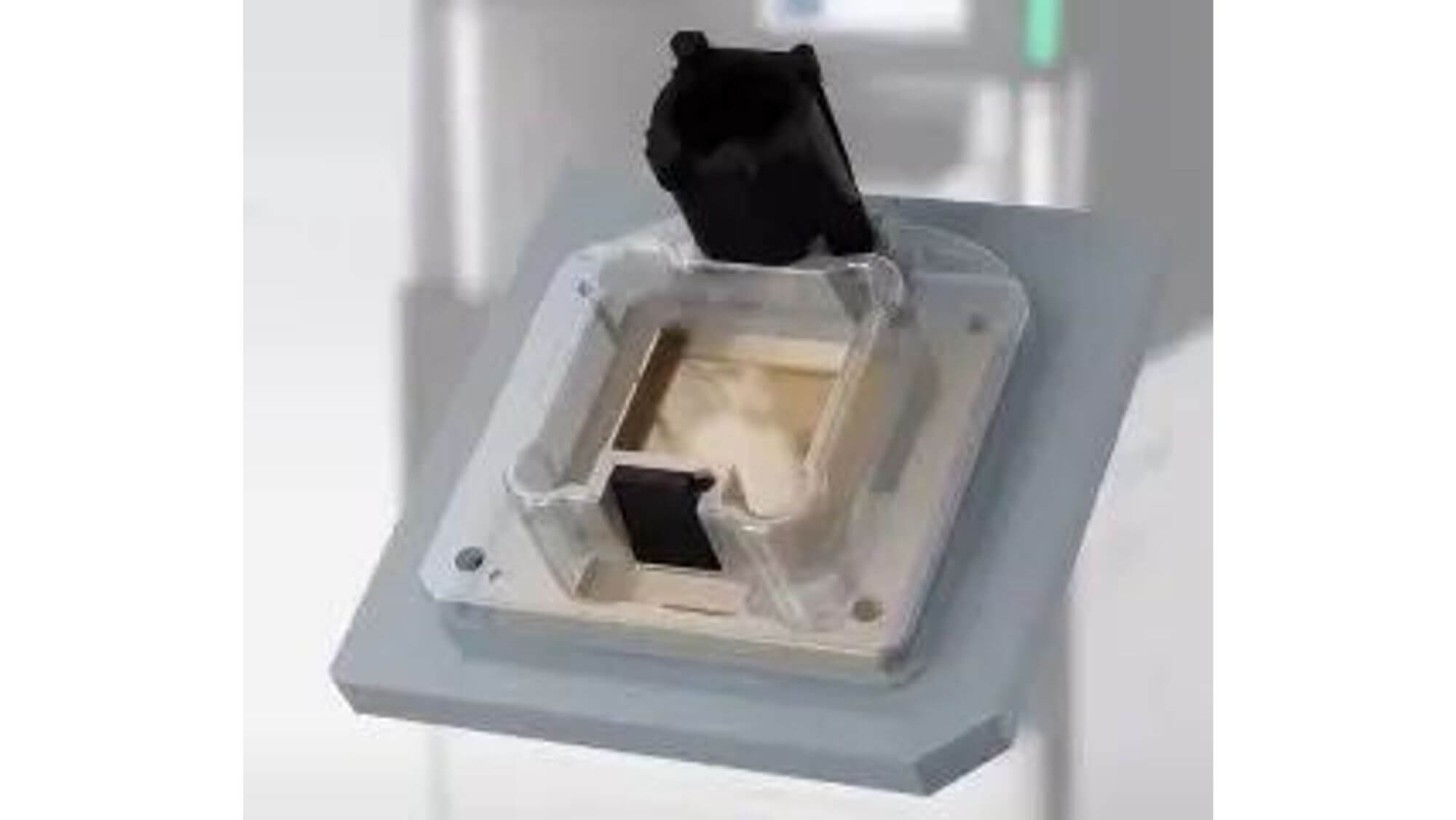
Zoomed in:

There are 150,000 sequencing micropores (Zero-Model Waveguides, ZMWs) neatly arranged on it, each with a diameter of 70 nanometers.
-
3. Construction of sequencing complex: polymerase, sequencing template, sequencing primer.
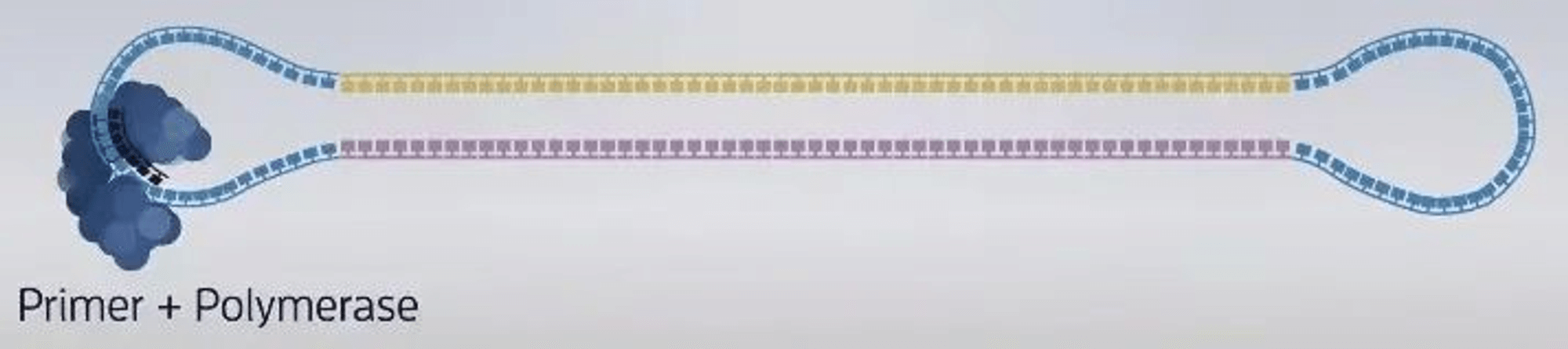
-
4. Scatter the complex into the sequencing pores.
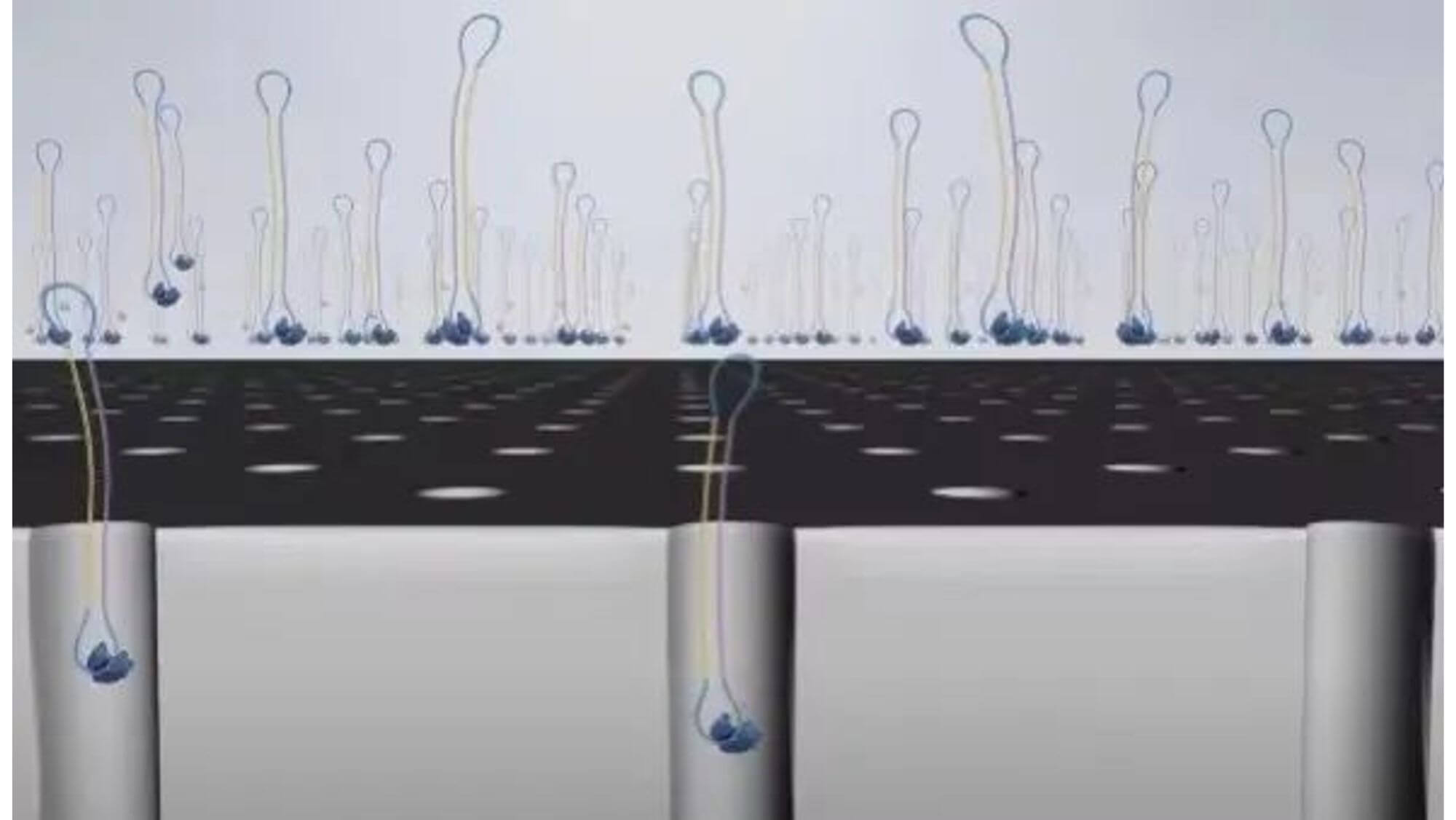
-
5. Since the polymerase is biotinylated, the glass substrate of the chip has streptavidin. Using the affinity of biotin and streptavidin, the sequencing complex containing polymerase will be fixed on the glass substrate.
-
6. The chip solution contains many free dNTPs, which are dNTPs randomly floating in the solution. The four bases of A, T, G, and C dNTP bear four corresponding colours of fluorescent groups on the phosphate group.
-
7. When synthesizing, the free dNTPs are captured by the enzyme fixed on the substrate, and lasers are emitted from the bottom of the glass plate.
Because the diameter of the sequencing micropore is very small, and the penetrability of the laser declines gradually, it can only transmit a short distance in the micropore. Therefore, only when the dNTP is close enough to the bottom, the fluorescent group will be irradiated by the laser and emit fluorescence.Of course, other free dNTPs may also float to the bottom of the micropore and be excited by light, but this situation is rare. Therefore, only one base will be measured at a time.After the synthesis of a base is completed, the fluorescently tagged phosphate group will fall off from the dNTP and undergo quenching, which does not affect the signal detection of other bases.
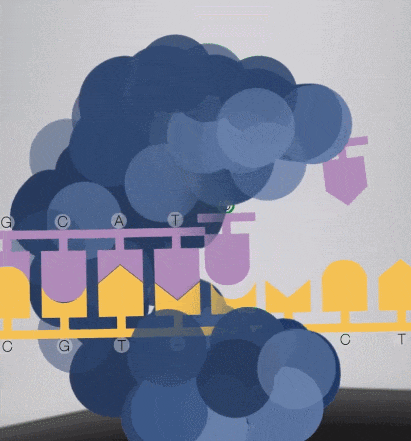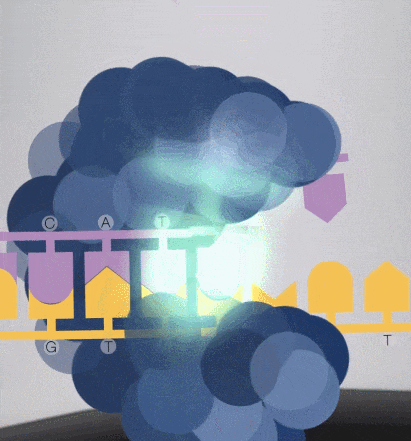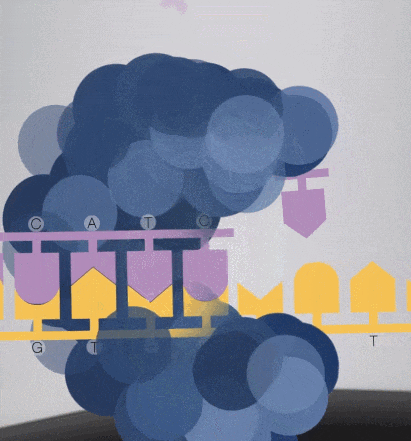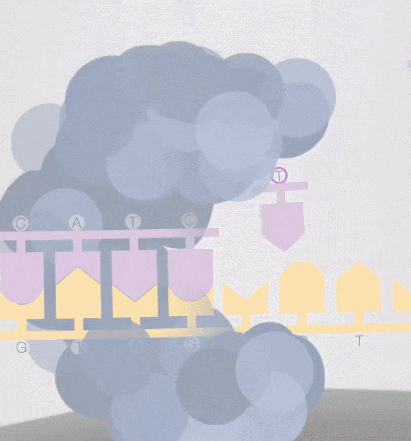
-
8. The sequencing pore where sequencing occurs has its own DNA fragments and sequencing complex, and different colors of excitation light are emitted at the same time.The machine will detect the following light signals, and in fact, up to tens of thousands of light points will be obtained at the same time.
-
9. Repeat the above steps, and after computer analysis of the spectrum, we finally get the sequencing files of the sample. In the SMRT sequencing process, about 10 bases are read per second, with a throughput of up to 7GB/day.
Interestingly, SMRT sequencing technology can directly detect the modified state of the bases during the sequencing process.For example, when the polymerase encounters a base with methylation, the synthesis speed will slow down significantly, and the spectrum will also change.
Therefore, SMRT sequencing technology can detect the methylation modification of the base.
Although the sequencing speed of SMRT sequencing technology is very fast, because it is single-molecule sequencing, each error generated in the reaction will be faithfully recorded, and it is difficult to distinguish. The sequencing accuracy rate is only 85%.
Fortunately, base reading errors are random, and if you read the same location again, the same error may not occur.If the same sequence is sequenced several times, these misread bases can be corrected. But compared to the accuracy rate of more than 99.5% of second-generation sequencing technology, this is indeed its biggest shortcoming.
Oxford Nanopore Technologies Nanopore Sequencing Technology
The concept of Nanopore sequencing was first proposed in the 1980s.
It is based on physical electronics and uses the change in local current when a single-strand DNA molecule passes through a nanopore to complete base sequence determination.
In 2005, Bayley established Oxford Nanopore Technologies (ONT) company. In 2014, the prototype of the first consumer-grade nanopore sequencer-MinION was born in ONT. It has attracted great attention from the scientific community since its release and is considered the most promising single-molecule sequencer.
Sequencing process:
- 1. DNA double strands unwind to form single-strand DNA.
- 2. At the same time, DNA helicase also acts as a motor protein to promote DNA single-strand molecules to pass through a biological nanopore constructed with α-hemolysin. The inner surface of the pore is covered with a synthetic cyclodextrin as a transducer.
- 3. DNA single strands stay in the pore, briefly interact with the cyclodextrin in the pore, affecting the original current flowing through the nanopore, bringing about current changes. Different bases bring different current changes. For example, the electric signal sizes of A and T are very close. However, T stays in the cyclodextrin for 2 to 3 times longer than other nucleotides, so each base is distinguished by their unique current interference amplitude.
- 4. Based on the spectrum of current changes, a pattern recognition algorithm is applied to get the base sequence.
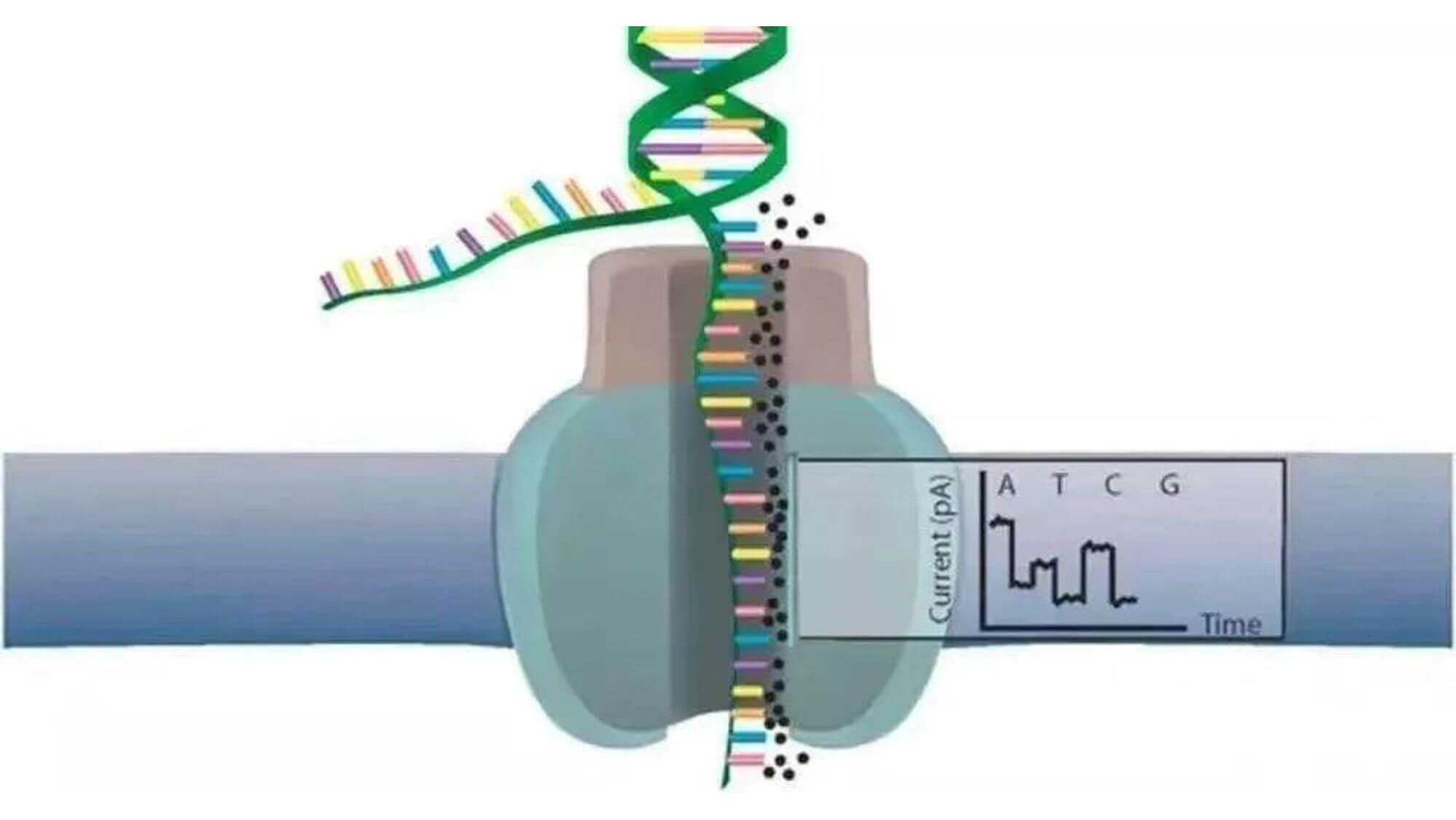
Main features:
-
1. Extra-long read length: In nanopore sequencing, the read length is not limited by the sequencing device and can be controlled by the library preparation experiment program used. The current record for the length of DNA fragments is up to 900kb.
-
2. Fast reading speed: MinION flow cell can read 500bp per second.
-
3. Direct sequencing: Nanopore technology is based on the principles of electronics, allowing direct sequencing of original DNA and RNA.
There is no need for DNA replication or chain synthesis, which saves time and cost. As nanopore technology supports direct sequencing without PCR, there is no amplification bias, and the library preparation workflow is also simpler.
-
4. High throughput: PromethION contains 48 independent flow cells and can output up to 2-4TB of data in 2 days.
-
5. Portable: ONT MinION is only the size of a USB device, also known as a palm sequencer, and can read data on a computer.
But at the same time, because this technology has over 1000 independent signals, its error rate is also higher (mainly manifested in the detection of Indel).
Since the modification of the base will change the originally set voltage change, the modification of the base is also a great challenge for ONT.
What is Indel:
In genome sequencing, "Indel" (insertion/deletion) refers to the variation of base insertion or deletion in the genome.
Insertion refers to the addition of one or more extra bases in the DNA sequence, while deletion refers to the deletion of one or more bases from the DNA sequence.These inserted or deleted bases can lead to changes in the sequence length in the genome, thereby affecting gene function.
Indel is one of the most common types of variations in the genome. Compared with a single base substitution (referred to as SNP), it usually has a greater impact on the gene function.
Indel can cause a reading frame shift, thereby changing the translation of the protein coding sequence, or causing functional changes in the non-coding region.
Therefore, for genome sequencing and genetic research, detecting and analyzing Indel mutations is very important and can help us understand the variations in the genome and their association with diseases.
Click to View → Mantacc Transport Mediums for gene sequencing






