News and Blogs
An Overall Comparison of PCR, RT-PCR, qPCR and RT-qPCR

Polymerase chain reaction (PCR) has revolutionized molecular biology and clinical diagnostics since its invention in the 1980s. As a versatile technique for amplification of DNA in vitro, PCR has spawned many variants that have extended its utility for diverse applications. Here we will explore the key differences between four common PCR-based techniques: basic PCR, reverse transcription PCR (RT-PCR), quantitative PCR (qPCR) and quantitative reverse transcription PCR (RT-qPCR).
What is Basic PCR?
The basic polymerase chain reaction exponentially amplifies a specific DNA target in a test tube. It utilizes heat-resistant DNA polymerases, primers complementary to the target sequence, nucleotides (dNTPs), and repetitive heating and cooling cycles to selectively amplify the target region of DNA. The result is millions to billions of copies of the specified DNA fragment. Applications of basic PCR include molecular cloning, mutation detection, pathogen identification, forensics, and paternity testing. It allows amplification of DNA extracted from any source for further qualitative or semi-quantitative analysis.
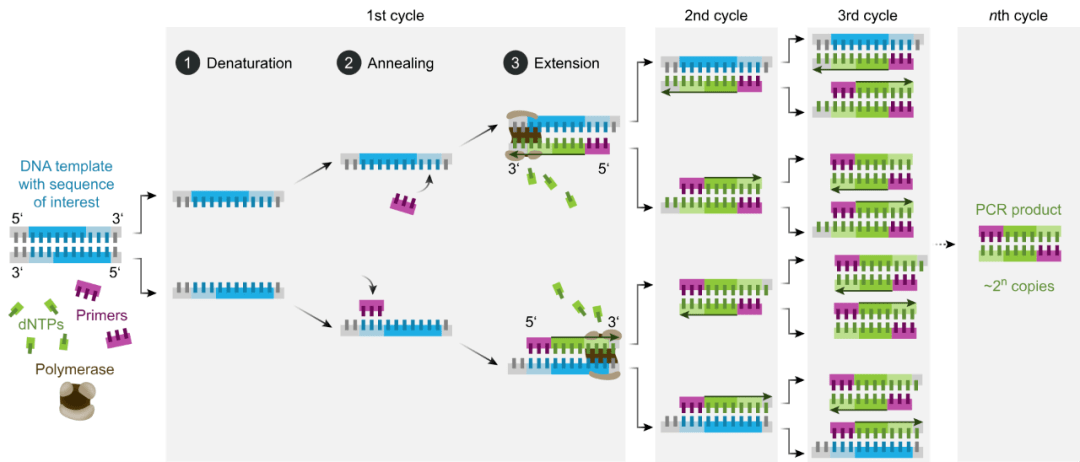
Principle of Basic PCR
A basic PCR consists of a series of 20-40 cycles. Each cycle has 3 discrete temperature steps:
- 1. Denaturation: The double-stranded DNA is heated to 94-98°C to separate it into two single-strands.
- 2. Annealing: The temperature is lowered to 50-65°C to allow primers to bind to their complementary sequences on the now single-stranded target DNA.
- 3. Extension: The temperature is raised to the ideal temperature for the DNA polymerase (around 72°C) and nucleotides are added to the primers for synthesis of new complementary strands along each single-stranded template.
After multiple cycles, the target region is exponentially amplified over a billion-fold. The amplicons can then be visualized using agarose gel electrophoresis.
What is RT-PCR?
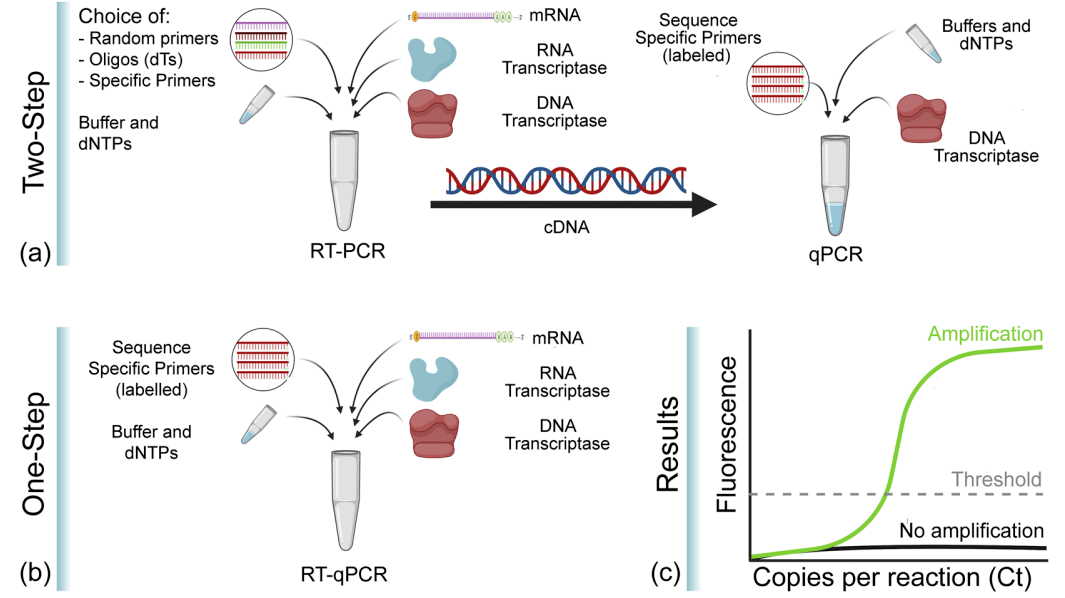
While basic PCR amplifies DNA, reverse transcription PCR (RT-PCR) allows amplification of RNA. First, an RNA template is converted into complementary DNA (cDNA) by a reverse transcriptase enzyme. Then this cDNA functions as the template for exponential amplification by regular PCR.
RT-PCR is useful for analyzing gene expression by amplifying mRNA transcripts present in small quantities. It is also used to detect and quantify RNA viruses. The cDNA amplicon can also be used for cloning RNA sequences.
RT-PCR can be one-step or two-step. In one-step RT-PCR, both reverse transcription and PCR occur sequentially in the same tube. In two-step RT-PCR, the cDNA synthesis reaction is performed separately before PCR amplification of the converted cDNA.
What is qPCR?
While basic PCR is primarily qualitative, quantitative PCR (qPCR) allows precise quantification of the amplified DNA. The amount of PCR product is measured at each cycle as amplification progresses. Monitoring amplification in real-time gives qPCR its common name – real-time PCR.
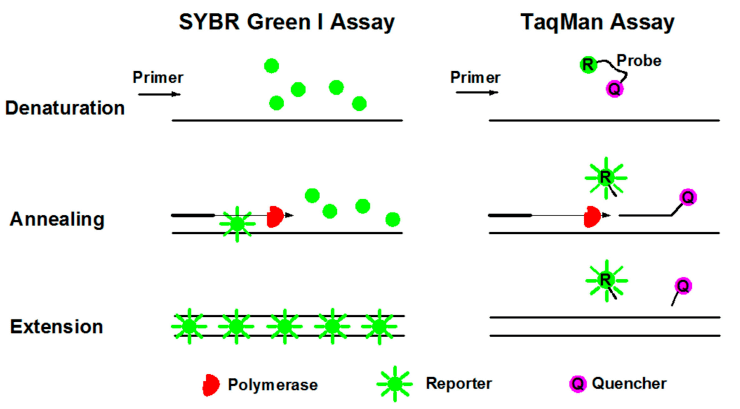
There are two main chemistries used for qPCR detection:
- 1. DNA binding dyes like SYBR Green - It gives fluorescence when bound to double-stranded DNA. So, fluorescent signal intensity reflects the accumulating PCR product with each cycle. But, lack of specificity can give false positive signals.
- 2. Sequence-specific probes like TaqMan - They produce a fluorescence signal only when bound to a complementary DNA target sequence. So, fluorescence intensity directly mirrors amplification of the intended target. Probes enhance specificity.
Standard curves help quantify nucleic acid concentration in unknown samples based on their PCR amplification profile compared to samples with a known concentration. qPCR allows highly accurate, sensitive and reproducible quantification of nucleic acids.
What is RT-qPCR?
RT-qPCR combines reverse transcription PCR with capabilities of qPCR for sensitive quantification of RNA. An RNA population is first converted to cDNA via reverse transcription, then amplified by real-time PCR with either DNA binding dyes or sequence-specific probes.
Uses and benefits of RT-qPCR:
- ✅Quantify gene expression by measuring RNA transcripts like mRNA
- ✅High specificity and accuracy
- ✅Rapid and reliable technique to analyze changes in gene expression
- ✅Diagnose and monitor infectious agents like HIV, influenza
- ✅Extreme sensitivity – can detect less than 5 copies
- ✅Cost-effective high throughput analysis
RT-qPCR vs qPCR vs RT-PCR vs Basic PCR
In summary:
- ✅Basic PCR amplifies DNA while RT-PCR amplifies RNA by first making cDNA via reverse transcription
- ✅qPCR quantifies DNA amplified in real-time while RT-qPCR quantifies RNA by converting it to cDNA before real-time amplification
- ✅Basic PCR and RT-PCR are qualitative or semi-quantitative while qPCR and RT-qPCR allow precise nucleic acid quantification
- ✅While RT-qPCR can reliably detect and measure tiny amounts of RNA, conventional PCR gives only qualitative yes/no detection for DNA.
| Method | Pros | Cons |
|---|---|---|
| PCR | Low cost; refined standards; products can be recycled for other experiments | Operation is cumbersome, specificity and sensitivity are low, prone to contamination, can only be qualitative but not quantitative. |
| qPCR | High specificity and sensitivity, easy operation: quantitative analysis can be performed. | High cost; products cannot be recycled. |
| RT-PCR and RT-qPCR | Applicable for all types of RNA. | RNA is prone to degradation: The RT step may increase the time and possibility of contamination. |
The myriad PCR technologies provide molecular biology laboratories with versatile tools for fundamental research and clinical diagnostics. Techniques like RT-qPCR pave the way for new frontiers like personalized medicine by enabling fast, affordable and accurate measurements of disease biomarkers.
Click to View high quality PCR consumables → Mantacc MBT-010 Viral Transport Medium (Non-inactivated)
References
- 1. Digital PCR as an Emerging Tool for Monitoring of Microbial Biodegradation.
- 2. A review on microscale polymerase chain reaction based methods in molecular diagnosis, and future prospects for the fabrication of fully integrated portable biomedical devices






